-
|
Hi, |
Beta Was this translation helpful? Give feedback.
Replies: 2 comments 5 replies
-
|
Oh yeah sorry, that is not well documented in the MICOM docs. Without QIIME you would use the Alternatively, you can use |
Beta Was this translation helpful? Give feedback.
-
|
Sorry, thought I already replied. The errors you are seeing are coming from multiprocessing (from the Python standard library) and have something to do how processes are spawned on your machine. Those are always hard to diagnose since they are often specific to the operating system and version and how you are executing your code. |
Beta Was this translation helpful? Give feedback.
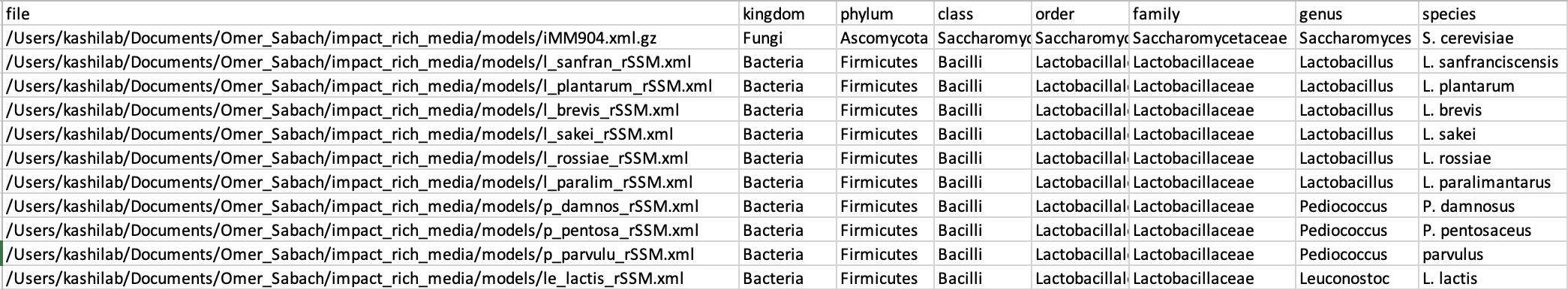
Oh yeah sorry, that is not well documented in the MICOM docs. Without QIIME you would use the
build_databasefunction. Theq2-micomtutorial has a bit more on that.Alternatively, you can use
buildwithout a model database. For that, your MICOM taxonomy table has to include a column namedfilethat points to a metabolic model in SBML format and that is consistent with the used taxonomic rank. Though, the recommendation would be to assemble a DB first. It makesbuildmuch faster, performs a lot of checks and fixes, and is easier to publish.